Int J Drug Res Clin. 2023;1:e10.
doi: 10.34172/ijdrc.2023.e10
Original Article
Blockade of CD155 and Interleukin-6 Suppresses Breast Cancer Cell Growth
Atefeh Khodakarami 1  , Samira Darvishi 2, Hadi Hassannia 3, Mohammad Sadeghi 4, Sana Esmaeeli 1, Vida Hashemi 5, Mohammad Hojjat-Farsangi 6, Farhad Jadidi-Niaragh 1, 7, *
, Samira Darvishi 2, Hadi Hassannia 3, Mohammad Sadeghi 4, Sana Esmaeeli 1, Vida Hashemi 5, Mohammad Hojjat-Farsangi 6, Farhad Jadidi-Niaragh 1, 7, * 
Author information:
1Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
2Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran
3Immunogenetic Research Center, Faculty of Medicine and Amol Faculty of Paramedical Sciences, Mazandaran University of Medical Sciences, Sari, Iran
4Research Center for Integrative Medicine in Aging, Aging Research Institute, Tabriz University of Medical Sciences, Tabriz, Iran
5Department of Basic Science, Faculty of Medicine, Maragheh University of Medical Sciences, Maragheh, Iran
6Bioclinicum, Department of Oncology-Pathology, Karolinska Institute, Stockholm, Sweden
7Department of Immunology, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
Abstract
Background:
Breast cancer is the foremost common type of cancer worldwide and the leading cause of death related to cancer. Poliovirus receptor (CD155) or nectin-like molecule-5 expression has been shown to increase in tumor cells and bind to the DNAX accessory molecule (DNAM-1)/T cell immunoreceptor with Ig and ITIM domains (TIGIT)/CD69 ligand present on the surface of T and natural killer cells, increasing inflammatory cytokines, including interleukin 6 (IL-6) in the tumor microenvironment. As a result, we intended to simultaneously target both CD155 and IL-6 as an effective therapeutic approach to prevent breast cancer growth.
Methods:
In this study, 4T1 cells were transfected with small interfering RNA (siRNA) molecules against CD155 and IL-6, using lipofectamine (LP). Target gene expression was analyzed using both the real-time polymerase chain reaction (RT-PCR) and an enzyme-linked immunosorbent assay (ELISA). The methyl thiazolyl tetrazolium (MTT) assay also examined the cytotoxic effect of combined therapy.
Results:
The results showed that the cells were effectively transfected with LP, resulting in the downregulation of IL-6 and CD155. In addition, the combined silencing of IL-6 and CD155 potently reduced the cellular viability of cancer cells.
Conclusion:
These findings revealed that the simultaneous targeting of CD155 and IL-6 in breast cancer cells (4T1) is an innovative and efficient therapy strategy to induce apoptosis in malignant cells.
Keywords: Breast cancer, CD155, Interleukin 6, 4T1, Apoptosis
Introduction
Breast cancer is a global malignancy that primarily affects females. Despite a positive response to treatment, resistance to therapy is common at advanced stages of this cancer.1,2 Surgery, chemotherapy, and radiotherapy are the primary treatment options that can be used to treat breast cancer. On the other hand, the new methods include antibody-drug conjugation systems, nanoparticle-based drug delivery systems, breast cancer stem cell-based therapies, and targeting prognostic and predictive biomarkers.3,4 The inefficiency of standard treatments in many patients has led many researchers to find new therapies.5,6 This low efficiency of common treatments is mainly due to the inhibitory microenvironment of the tumor.7 The tumor microenvironment contains a variety of immunosuppressive mechanisms which coordinate the suppression of anti-tumor immune responses.8,9 Therefore, one way to inhibit tumor growth is by inhibiting or preventing the function of these inhibitory mechanisms in the tumor microenvironment. Of the different mechanisms that inhibit immune responses in the tumor microenvironment, the CD155/ interleukin 6) IL-6( axis plays a crucial role in tumor progression.
The poliovirus receptor, also known as nectin-like molecule-5, or CD155 (Transmembrane glycoprotein type I receptor belongs to the immunoglobulin superfamily) acts as a protein for immune checkpoint. It alters the immune response in the function of the tumor microenvironment.10,11 Since CD155 contains immune-receptor tyrosine-based inhibitory motifs (ITIM), it activates T/NK by binding to DNAX accessory molecule (DNAM-1) but inhibits immune responses when attached to T cell immunoreceptor with Ig and ITIM domains/CD69.11,12 The inhibitory function of CD155 in the tumor microenvironment is more significant than its activation function.13 Furthermore, CD155 expression is high in many tumors and a poor prognostic factor.14,15 The overexpression of CD155 amplifies tumor cell migration, metastasis, growth, and resistance to therapy.16-18
Moreover, CD155 promotes the expression of IL-6/ transforming growth factor beta / SMAD family member 3 by being directly associated with each of these factors.10,19 In addition, the increased expression of IL-6 in breast cancer promotes metastasis, migration, and angiogenesis.20,21 IL-6 plays a crucial role in cancer, and patients whose serum levels of IL-6 are high have a low rate of recovery and survival; on the contrary, the rate of recovery is higher in patients with low IL-6 serum levels22; this cytokine is extensively secreted by both immune cells and cancer cells in the tumor microenvironment.23 Signaling through a signal transducer and activator of transcription 3 (STAT3) inhibits the activity of immune cells against tumor cells.24 However, it also stimulates cancer cell growth, angiogenesis, metastasis, and resistance to chemotherapy.25
Therefore, we hypothesized that combined methods for targeting these factors would result in an effective method for cancer therapy. It also inhibits the growth of tumor cells and induces apoptosis, making it a promising combination treatment for breast cancer.
Methods
Reagents
CD155 and IL-6 siRNA molecules and small interfering RNA (siRNA) negative control (NC-siRNA) have been bought from Santa Cruz Biotechnology, Inc (CA, USA). The methyl thiazolyl tetrazolium (MTT) cell proliferation assay kit was purchased from Sigma-Aldrich (Bornem, Belgium) and used according to the guidelines supplied by the company.
Cell Culture
4T1 cells (ER- PR- HER2-) were grown in RPMI 1640 (Gibco, US). In the culture medium, 10% fetal bovine serum was added. All cells were cultivated in a 37°C humidified incubator using 5% CO2.
Method of Cell Transfections and Evaluation of Transfection Efficiency
After seeding 1 × 104 cells/wells in a 96-well plate and incubating at 37°C for 24 hours, cells were transfected with 90 pm siRNA via Lipofectamine (LP) RNAiMax (Invitrogen, Karlsruhe, Germany) in accordance with the company’s instructions. To analyze transfection efficiency, a cellular uptake assay was performed. In brief, cells were transfected with PE-labeled siRNAs encapsulated in LP, and the frequency of cells transfected with siRNA was measured using flow cytometry assay.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from 4T1 cells by TRIzol Reagent (TaKaRa) based on the manufacturer’s instructions. For reverse transcription,2 μg of total RNA was used in a final volume of 20 μL. Table 1 shows the primer sequences.17,21,26 Quantitative real-time polymerase chain reaction (qRT-PCR) was done by 2x SYBR Green Mix (Promega, Madison, USA) using Roche Light Cycler 480 PCR cycler. Then, fold change was assessed using the formulation 2-ΔΔCt, and each reaction was carried out in triplicate.
Table 1.
Primer Sequences
|
Primer name
|
|
Sequences
|
| B-actin |
Forward |
5′- GGTCATCACTATTGGCAACG-3′ |
| Reverse |
5′- ACGGATGTCAACGTCACACT-3′ |
| CD155 |
Forward |
5′- GCTAGAAGGACTCACTAGACTCAGGAA-3′ |
| Reverse |
5′- GTCGCCTCATCTGTCGTGGAAC-3′ |
| IL-6 |
Forward |
5′- ATCCAGTTGCCTTCTTGGGACTGA -3′ |
| Reverse |
5′- TAAGCCTCCGACTTGTGAAGTGGT -3′ |
| Bax |
Forward |
5′ - GCCGAAATGTTTGCTGACG -3′ |
| Reverse |
5′ - CAGCCGATCTCGAAGGAAG -3′ |
| Bcl2 |
Forward |
5′ - GGCTGGGGATGACTTCTCTC -3′ |
| Reverse |
5′ - ACAATCCTCCCCCAGTTCAC -3′ |
| STAT3 |
Forward |
5′ - CACGAAAGTCAGGTTGCTGGT -3′ |
| Reverse |
5′ - ACTTTTGTGTTCGTGCCCAGA -3′ |
| Caspase-3 |
Forward |
5′ - TGGGACTGATGAGGAGATGG -3′ |
| Reverse |
5′ - CTGCAAAGGGACTGGATGAA -3′ |
Note. IL-6: Interleukin 6; STAT3: Signal transducer and activator of transcription 3.
Cytotoxicity Assay
MTT test was done on 4T1 cell lines after 48 hours of incubation with siRNA. Then, 1 × 104 cells were seeded in 96-well plates to analyze the cytotoxicity of siRNA-loaded LP. Moreover, 4T1 cells were incubated with CD155 siRNA-loaded LPs (90 pm siRNA), IL-6 siRNA-loaded LPs, and IL-6-CD155 siRNA-loaded LPs, and their viability was assessed relative to blank LPs. Then, 20 μL of MTT solution was added to each well where 150 µL of dimethyl sulfoxide was used to replace the supernatant after four hours of exposure. Finally, after 30 minutes, the OD of the wells was assessed using a spectrophotometer (Synergy 4, BioTec, USA), and the test was performed in triplicate.
Cytokine Release Assay by an Enzyme-Linked Immunosorbent Assay
To evaluate the levels of IL-6 cytokine secretion from cancer cells, 8 × 104 4T1 cells were seeded in 12-well plates and incubated with siRNA for 48 hours. Then, IL-6 secretion levels were measured using an enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen, Carlsbad, CA, USA).
Cell Proliferation Analysis by Bromodeoxyuridine
The proliferation of tumor cells was assessed using a cell proliferation ELISA Bromodeoxyuridine (BrdU) Kit (Roche’s, Mannheim, Germany). Afterward, 1 × 104 tumor cells were cultured for 24 hours in a 96-well plate at 37°C in 5% CO2. Each well was then incubated for 48 hours at 37°C with siRNA-loaded LPs added. After that, the fresh culture medium was added, and cells were incubated for 24 hours at 37°C with 5% CO2. After filling each well with 10 Mm BrdU labeling solution, incubation lasted for 24 hours. After cell fixation, a FixDenat solution was used to denature the DNA of tumor cells. Following that, an anti-BrdU-POD solution was applied to each well. The immune complexes were then detected using a microplate luminometer (LuminoskanTM Microplate Luminometer, Thermo Fisher Scientific, USA) by adding a luminescence substrate solution to each well.
Statistical Analysis
GraphPad Prism V9 software and a two-way ANOVA test were used for statistical analysis, and statistical significance was set at P < 0.05.
Results
Lipofectamine Effectively Transfected Small Interfering RNA into Cells and Silenced the Expression of Target Cells
A flow cytometry assay was used to measure how quickly cancer cells were transfected with siRNA using LP and how effectively it did so. As shown in Figure 1a, about 70% of cells were successfully uptaking siRNA molecules encapsulated in LP. Using qRT-PCR, the levels of CD155-IL-6 mRNA in cells treated with various LP treatment groups were evaluated to determine how well gene silencing worked. Figure 1b-c indicates the outcomes of changes in CD155-IL-6 mRNA levels in other treatment groups. The results revealed no changes in cells treated with scramble siRNA or LP alone. Furthermore, there were no significant changes in mRNA levels in cells treated with CD155 or IL-6 siRNA independently, indicating that a carrier is required for effective siRNA delivery into cells. However, the combined treatment of cells with CD155-IL-6 siRNA-LP efficiently reduced CD155/IL-6 mRNA expression.
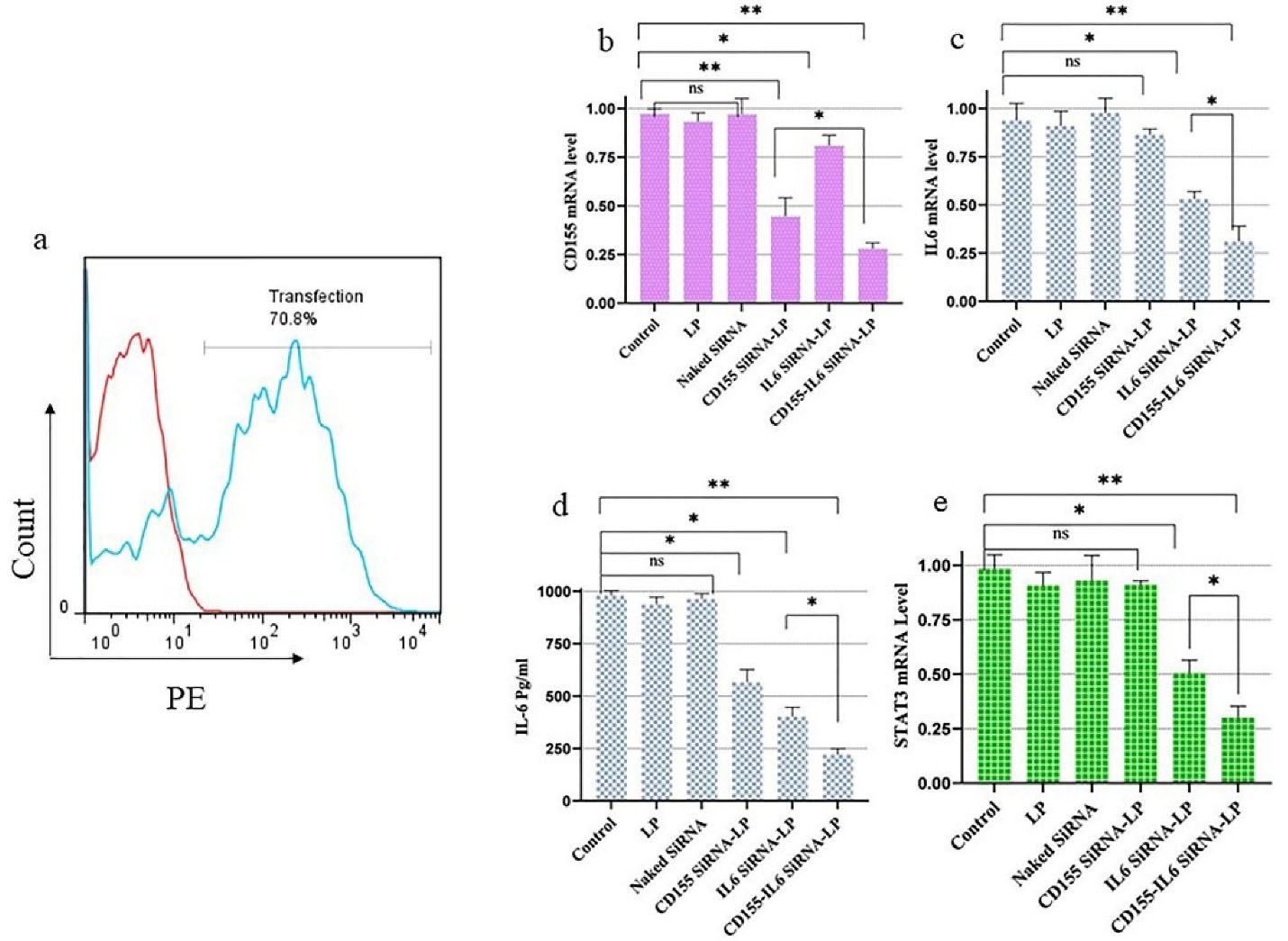
Figure 1.
Cancer Cells Were Effectively Transfected by siRNA Molecules Loaded in LP. (a) Transfection efficiency was measured using flow cytometry using PE-labeled siRNA molecules loaded in LP. (b) The transfection of 4T1 cells with LP-CD155 siRNA suppressed the expression of CD155. (c) The treatment of 4T1 cells with anti-IL-6 siRNA using LP led to suppressing the IL-6 expression as illustrated by the real-time assay. (d) The protein level of IL-6 release is measured by ELISA. (e) STAT3 expression decreased through IL-6 blockage. Note. siRNA: Small interfering RNA; LP: Lipofectamine; IL-6: Interleukin 6; ELISA: Enzyme-linked immunosorbent assay; STAT3: Signal transducer and activator of transcription 3; *P < 0.05; ** P < 0.01, ns: Non-significant
.
Cancer Cells Were Effectively Transfected by siRNA Molecules Loaded in LP. (a) Transfection efficiency was measured using flow cytometry using PE-labeled siRNA molecules loaded in LP. (b) The transfection of 4T1 cells with LP-CD155 siRNA suppressed the expression of CD155. (c) The treatment of 4T1 cells with anti-IL-6 siRNA using LP led to suppressing the IL-6 expression as illustrated by the real-time assay. (d) The protein level of IL-6 release is measured by ELISA. (e) STAT3 expression decreased through IL-6 blockage. Note. siRNA: Small interfering RNA; LP: Lipofectamine; IL-6: Interleukin 6; ELISA: Enzyme-linked immunosorbent assay; STAT3: Signal transducer and activator of transcription 3; *P < 0.05; ** P < 0.01, ns: Non-significant
ELISA analysis confirmed the decrease in IL-6 secretion to examine the effect of treatment on IL-6 protein levels. Compared to untreated groups, the secretin levels of IL-6 markedly decreased in mono- and combination-treated groups.
Furthermore, STAT3 expression in the study groups was measured to find out how IL-6 inhibition affected the STAT3 signaling factor. In the groups treated with CD155-IL-6 siRNA-LP and IL-6 siRNA-LP, STAT3 expression was significantly reduced; however, CD155 inhibition did not affect STAT3 expression (Figure 1e).
CD155-IL-6 Inhibition Induces Apoptosis and Inhibits Proliferation in 4T1 Cells
The viability of 4T1 cells after 24 or 48 hours of incubation was examined using an MTT test to assess the impact of various treatments. Figures 2a-b present the findings. The viability of controls, including untreated and LP, did not change significantly. In contrast, cells treated with CD155 or IL-6 siRNA alone reduced viability considerably. Furthermore, compared to controls, the apoptosis rate in the CD155-IL-6 siRNA-LP-treated group was significantly higher.
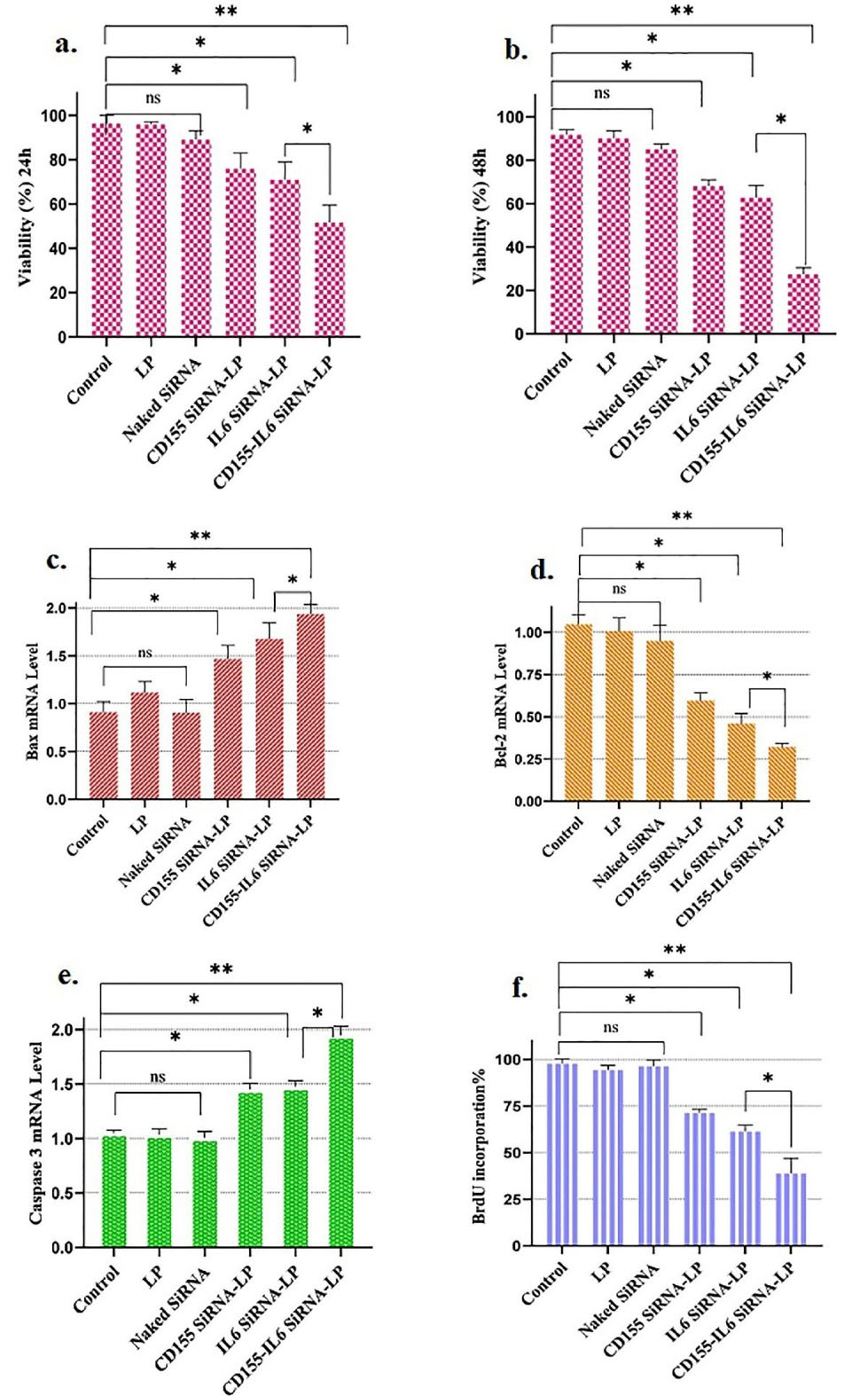
Figure 2.
Silencing CD155/IL-6 Decreased the Viability of 4T1 Cells. (a-b) 4T1 cells were treated with the combination of anti-CD155-IL-6 siRNA-LP-, and the toxic effect was analyzed using MTT assay after 24 and 48 hours of incubation. (c) Bax expression level enhanced through combined treatment. (d) Bcl2 mRNA level diminished by combined treatment. (e) Caspase-3 expression level enhanced through combined treatment. (f) BrdU analysis of cell proliferation. Note. IL-6: Interleukin 6; siRNA: Small interfering RNA; LP: Lipofectamine; IL-6: Interleukin 6; MTT: Methyl thiazolyl tetrazolium; * P < 0.05; ** P < 0.01; ns: Non-significant
.
Silencing CD155/IL-6 Decreased the Viability of 4T1 Cells. (a-b) 4T1 cells were treated with the combination of anti-CD155-IL-6 siRNA-LP-, and the toxic effect was analyzed using MTT assay after 24 and 48 hours of incubation. (c) Bax expression level enhanced through combined treatment. (d) Bcl2 mRNA level diminished by combined treatment. (e) Caspase-3 expression level enhanced through combined treatment. (f) BrdU analysis of cell proliferation. Note. IL-6: Interleukin 6; siRNA: Small interfering RNA; LP: Lipofectamine; IL-6: Interleukin 6; MTT: Methyl thiazolyl tetrazolium; * P < 0.05; ** P < 0.01; ns: Non-significant
To confirm apoptosis further, the expression of Bax, Bcl2, and Caspase-3, which are involved in cell apoptosis, was measured. The expression of the pro-apoptotic factors Bax and Caspase-3 increased in groups that received CD155 siRNA-LP or IL-6 siRNA-LP alone, while the expression of the anti-apoptotic factor Bcl2 decreased (Figure 2c-e). In addition, the group that received the combined treatment exhibited higher mRNA levels of Bax and caspase-3 and a lower Bcl2 mRNA level (Figure 2c-e). The BrdU assay kit showed that the simultaneous inhibition of CD155 and IL-6 in the CD155-IL-6 siRNA-LP study group reduced the proliferation of 4T1 cancer cells (Figure 2f).
Discussion
Changing the tumor microenvironment is one of the most critical activities that immunotherapists can do to treat cancer. To evade drugs and immune responses, tumor cells modify themselves through a variety of mechanisms, including immunosuppressive cells, inhibitory cytokines, checkpoints, hypoxia, and metabolic changes.27 The immune checkpoint targeting offers a significant clinical opportunity for solid tumor patients.28 Various inhibitory checkpoints are expressed in the tumor microenvironment, contributing to tumor growth and the inhibition of immune responses. CD155, as an immunomodulatory factor, has an inhibitory function in the immune microenvironment of breast cancer.29 CD155 induced a higher proportion of exhausted CD4+ and unexhausted CD8+ TILs and a higher PD-L1/TIGIT expression on immune cells.2,30,31 Therefore, we hypothesized that CD155 targeting could be an effective agent in breast cancer therapy. In this regard, the suppression of CD155, an immunomodulatory factor, has been shown to be a useful agent for targeting solid tumors in several studies.28,32 Moreover, the researchers found that the methylation at the CpG site of the CD155 promoter is directly related to cancer progression and tumor size.33 However, there is little information about the role of this biomarker (CD155) in breast cancer.34 Our findings revealed that the expression of this biomarker increases in breast cancer.
In addition, in line with other studies,10,19 the present study found that 4T1 cells increase the expression of CD155 and IL-6. Moreover, this study showed the role of IL-6 in the microenvironment of breast cancer in several studies.21,26,35 Treatments that target IL-6, IL-6R, glycoprotein 130 receptor, Janus kinase, and STAT3, which form an interconnected loop, have been shown to reduce tumor resistance in some studies.36,37 Avalle et al discovered that in the microenvironment of breast tumor cells, cancer-associated fibroblasts exhibit an elevated expression of STAT3, which was activated by IL 6, and that STAT3 boosts tumor cell proliferation and invasion.38
In general, we found that targeting two biomarkers IL-6/CD155 together, considering the role of each factor in breast cancer,2,26,30 can be a promising combined treatment. In this study, for the first time, we silenced both of these biomarkers simultaneously and looked at how the treatment affected cancer cells. We used siRNA molecules to inhibit CD155/IL-6 expression, but since siRNA molecules are highly unstable and susceptible to degradation by nuclease enzymes,39,40 we needed a suitable carrier to inhibit CD155/IL-6 expression. We used LP for siRNA transfection due to its low toxicity and effective cargo delivery.41 As a consequence of this, the malignant cell survival rate decreases with the combined treatment that we assumed to be effective. Accordingly, it is suggested as a promising treatment method in future studies.
Ethics statement
The Ethics Committee of Tabriz University of Medical Sciences has approved this study.
Disclosure of funding source
Financial support for this study was by the Tabriz University of Medical Sciences (grant number: 69959).
Conflict of interests declaration
None.
Data availability statement
The data supporting this study’s findings are available on request from the corresponding author following the reasonable request.
Consent for publication
Not applicable.
References
- Katsura C, Ogunmwonyi I, Kankam HK, Saha S. Breast cancer: presentation, investigation and management. Br J Hosp Med (Lond) 2022; 83(2):1-7. doi: 10.12968/hmed.2021.0459 [Crossref] [ Google Scholar]
- Wang RB, Li YC, Zhou Q, Lv SZ, Yuan KY, Wu JP. Overexpression of CD155 is associated with PD-1 and PD-L1 expression on immune cells, rather than tumor cells in the breast cancer microenvironment. World J Clin Cases 2020; 8(23):5935-43. doi: 10.12998/wjcc.v8.i23.5935 [Crossref] [ Google Scholar]
- Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S. Breast cancer: biology, biomarkers, and treatments. Int Immunopharmacol 2020; 84:106535. doi: 10.1016/j.intimp.2020.106535 [Crossref] [ Google Scholar]
- Peart O. Breast intervention and breast cancer treatment options. Radiol Technol 2015; 86(5):535M-58M. [ Google Scholar]
- Arruebo M, Vilaboa N, Sáez-Gutierrez B, Lambea J, Tres A, Valladares M. Assessment of the evolution of cancer treatment therapies. Cancers (Basel) 2011; 3(3):3279-330. doi: 10.3390/cancers3033279 [Crossref] [ Google Scholar]
- Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol 2005; 205(2):275-92. doi: 10.1002/path.1706 [Crossref] [ Google Scholar]
- Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer 2019; 18(1):10. doi: 10.1186/s12943-018-0928-4 [Crossref] [ Google Scholar]
- Mhaidly R, Mechta-Grigoriou F. Fibroblast heterogeneity in tumor micro-environment: role in immunosuppression and new therapies. Semin Immunol 2020; 48:101417. doi: 10.1016/j.smim.2020.101417 [Crossref] [ Google Scholar]
- Shimizu K, Iyoda T, Okada M, Yamasaki S, Fujii SI. Immune suppression and reversal of the suppressive tumor microenvironment. Int Immunol 2018; 30(10):445-54. doi: 10.1093/intimm/dxy042 [Crossref] [ Google Scholar]
- Chashchina A, Märklin M, Hinterleitner C, Salih HR, Heitmann JS, Klimovich B. DNAM-1/CD226 is functionally expressed on acute myeloid leukemia (AML) cells and is associated with favorable prognosis. Sci Rep 2021; 11(1):18012. doi: 10.1038/s41598-021-97400-6 [Crossref] [ Google Scholar]
- Yoshikawa K, Ishida M, Yanai H, Tsuta K, Sekimoto M, Sugie T. Immunohistochemical analysis of CD155 expression in triple-negative breast cancer patients. PLoS One 2021; 16(6):e0253176. doi: 10.1371/journal.pone.0253176 [Crossref] [ Google Scholar]
- Oda T, Ohka S, Nomoto A. Ligand stimulation of CD155alpha inhibits cell adhesion and enhances cell migration in fibroblasts. Biochem Biophys Res Commun 2004; 319(4):1253-64. doi: 10.1016/j.bbrc.2004.05.111 [Crossref] [ Google Scholar]
- Kučan Brlić P, Lenac Roviš T, Cinamon G, Tsukerman P, Mandelboim O, Jonjić S. Targeting PVR (CD155) and its receptors in anti-tumor therapy. Cell Mol Immunol 2019; 16(1):40-52. doi: 10.1038/s41423-018-0168-y [Crossref] [ Google Scholar]
- González-Ochoa S, Tellez-Bañuelos MC, Méndez-Clemente AS, Bravo-Cuellar A, Hernández Flores G, Palafox-Mariscal LA. Combination blockade of the IL-6R/STAT-3 axis with TIGIT and Its impact on the functional activity of NK cells against prostate cancer cells. J Immunol Res 2022; 2022:1810804. doi: 10.1155/2022/1810804 [Crossref] [ Google Scholar]
- Triki H, Charfi S, Bouzidi L, Ben Kridis W, Daoud J, Chaabane K. CD155 expression in human breast cancer: clinical significance and relevance to natural killer cell infiltration. Life Sci 2019; 231:116543. doi: 10.1016/j.lfs.2019.116543 [Crossref] [ Google Scholar]
- Boissière-Michot F, Chateau MC, Thézenas S, Guiu S, Bobrie A, Jacot W. Correlation of the TIGIT-PVR immune checkpoint axis with clinicopathological features in triple-negative breast cancer. Front Immunol 2022; 13:1058424. doi: 10.3389/fimmu.2022.1058424 [Crossref] [ Google Scholar]
- Gao J, Zheng Q, Shao Y, Wang W, Zhao C. CD155 downregulation synergizes with adriamycin to induce breast cancer cell apoptosis. Apoptosis 2018; 23(9-10):512-20. doi: 10.1007/s10495-018-1473-8 [Crossref] [ Google Scholar]
- Zhang D, Liu J, Zheng M, Meng C, Liao J. Prognostic and clinicopathological significance of CD155 expression in cancer patients: a meta-analysis. World J Surg Oncol 2022; 20(1):351. doi: 10.1186/s12957-022-02813-w [Crossref] [ Google Scholar]
- Zheng Q, Gao J, Yin P, Wang W, Wang B, Li Y. CD155 contributes to the mesenchymal phenotype of triple-negative breast cancer. Cancer Sci 2020; 111(2):383-94. doi: 10.1111/cas.14276 [Crossref] [ Google Scholar]
- Martínez-Pérez C, Kay C, Meehan J, Gray M, Dixon JM, Turnbull AK. The IL-6-like cytokine family: role and biomarker potential in breast cancer. J Pers Med 2021; 11(11):1073. doi: 10.3390/jpm11111073 [Crossref] [ Google Scholar]
- Masjedi A, Ahmadi A, Atyabi F, Farhadi S, Irandoust M, Khazaei-Poul Y. Silencing of IL-6 and STAT3 by siRNA loaded hyaluronate-N,N,N-trimethyl chitosan nanoparticles potently reduces cancer cell progression. Int J Biol Macromol 2020; 149:487-500. doi: 10.1016/j.ijbiomac.2020.01.273 [Crossref] [ Google Scholar]
- Chen MF, Chen PT, Lu MS, Lin PY, Chen WC, Lee KD. IL-6 expression predicts treatment response and outcome in squamous cell carcinoma of the esophagus. Mol Cancer 2013; 12:26. doi: 10.1186/1476-4598-12-26 [Crossref] [ Google Scholar]
- Wang J, Li D, Cang H, Guo B. Crosstalk between cancer and immune cells: Role of tumor-associated macrophages in the tumor microenvironment. Cancer Med 2019; 8(10):4709-21. doi: 10.1002/cam4.2327 [Crossref] [ Google Scholar]
- Wang Y, Shen Y, Wang S, Shen Q, Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett 2018; 415:117-28. doi: 10.1016/j.canlet.2017.12.003 [Crossref] [ Google Scholar]
- Browning L, Patel MR, Horvath EB, Tawara K, Jorcyk CL. IL-6 and ovarian cancer: inflammatory cytokines in promotion of metastasis. Cancer Manag Res 2018; 10:6685-93. doi: 10.2147/cmar.s179189 [Crossref] [ Google Scholar]
- Salimifard S, Karoon Kiani F, Sadat Eshaghi F, Izadi S, Shahdadnejad K, Masjedi A. Codelivery of BV6 and anti-IL-6 siRNA by hyaluronate-conjugated PEG-chitosan-lactate nanoparticles inhibits tumor progression. Life Sci 2020; 260:118423. doi: 10.1016/j.lfs.2020.118423 [Crossref] [ Google Scholar]
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015; 348(6230):56-61. doi: 10.1126/science.aaa8172 [Crossref] [ Google Scholar]
- Fang J, Chen F, Liu D, Gu F, Chen Z, Wang Y. Prognostic value of immune checkpoint molecules in breast cancer. Biosci Rep 2020; 40(7):BSR20201054. doi: 10.1042/bsr20201054 [Crossref] [ Google Scholar]
- Briukhovetska D, Suarez-Gosalvez J, Voigt C, Markota A, Giannou AD, Schübel M, et al. T cell-derived interleukin-22 drives the expression of CD155 by cancer cells to suppress NK cell function and promote metastasis. Immunity 2023;56(1):143-61.e11. 10.1016/j.immuni.2022.12.010.
- Chen C, Guo Q, Fu H, Yu J, Wang L, Sun Y. Asynchronous blockade of PD-L1 and CD155 by polymeric nanoparticles inhibits triple-negative breast cancer progression and metastasis. Biomaterials 2021; 275:120988. doi: 10.1016/j.biomaterials.2021.120988 [Crossref] [ Google Scholar]
- Li YC, Zhou Q, Song QK, Wang RB, Lyu S, Guan X. Overexpression of an immune checkpoint (CD155) in breast cancer associated with prognostic significance and exhausted tumor-infiltrating lymphocytes: a cohort study. J Immunol Res 2020; 2020:3948928. doi: 10.1155/2020/3948928 [Crossref] [ Google Scholar]
- Iguchi-Manaka A, Okumura G, Ichioka E, Kiyomatsu H, Ikeda T, Bando H. High expression of soluble CD155 in estrogen receptor-negative breast cancer. Breast Cancer 2020; 27(1):92-9. doi: 10.1007/s12282-019-00999-8 [Crossref] [ Google Scholar]
- Triki H, Declerck K, Charfi S, Ben Kridis W, Chaabane K, Ben Halima S. Immune checkpoint CD155 promoter methylation profiling reveals cancer-associated behaviors within breast neoplasia. Cancer Immunol Immunother 2022; 71(5):1139-55. doi: 10.1007/s00262-021-03064-6 [Crossref] [ Google Scholar]
- Yong H, Cheng R, Li X, Gao G, Jiang X, Cheng H. CD155 expression and its prognostic value in postoperative patients with breast cancer. Biomed Pharmacother 2019; 115:108884. doi: 10.1016/j.biopha.2019.108884 [Crossref] [ Google Scholar]
- Masjedi A, Hashemi V, Hojjat-Farsangi M, Ghalamfarsa G, Azizi G, Yousefi M. The significant role of interleukin-6 and its signaling pathway in the immunopathogenesis and treatment of breast cancer. Biomed Pharmacother 2018; 108:1415-24. doi: 10.1016/j.biopha.2018.09.177 [Crossref] [ Google Scholar]
- Manore SG, Doheny DL, Wong GL, Lo HW. IL-6/JAK/STAT3 signaling in breast cancer metastasis: biology and treatment. Front Oncol 2022; 12:866014. doi: 10.3389/fonc.2022.866014 [Crossref] [ Google Scholar]
- Siersbæk R, Scabia V, Nagarajan S, Chernukhin I, Papachristou EK, Broome R, et al. IL-6/STAT3 signaling hijacks estrogen receptor α enhancers to drive breast cancer metastasis. Cancer Cell 2020;38(3):412-23.e9. 10.1016/j.ccell.2020.06.007.
- Avalle L, Raggi L, Monteleone E, Savino A, Viavattene D, Statello L. STAT3 induces breast cancer growth via ANGPTL4, MMP13 and STC1 secretion by cancer associated fibroblasts. Oncogene 2022; 41(10):1456-67. doi: 10.1038/s41388-021-02172-y [Crossref] [ Google Scholar]
- Higuchi Y, Kawakami S, Hashida M. Strategies for in vivo delivery of siRNAs: recent progress. BioDrugs 2010; 24(3):195-205. doi: 10.2165/11534450-000000000-00000 [Crossref] [ Google Scholar]
- Sajid MI, Moazzam M, Kato S, Yeseom Cho K, Tiwari RK. Overcoming barriers for siRNA therapeutics: from bench to bedside. Pharmaceuticals (Basel) 2020; 13(10):294. doi: 10.3390/ph13100294 [Crossref] [ Google Scholar]
- Yu X, Liang X, Xie H, Kumar S, Ravinder N, Potter J. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol Lett 2016; 38(6):919-29. doi: 10.1007/s10529-016-2064-9 [Crossref] [ Google Scholar]